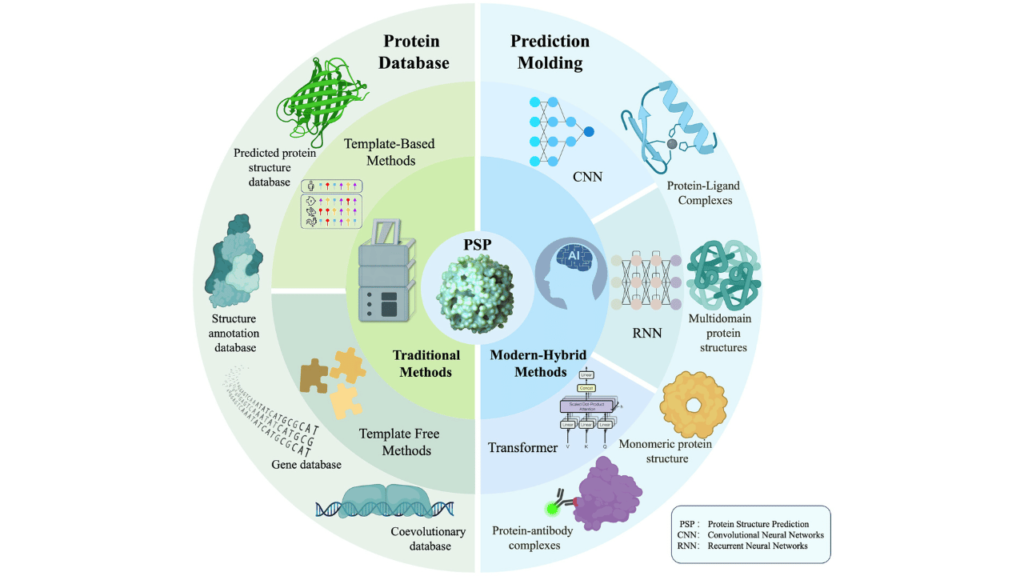
Proteins are the building blocks of life, yet classifying them into families remains one of the greatest challenges in computational biology. With over 200 million protein sequences cataloged and more being discovered every day, how do we make sense of this vast biological data? Traditional methods struggle with novel or low-sample proteins. Enter deep few-shot learning—a game-changing approach that redefines how we identify and categorize protein families with minimal data.
Few-shot learning isn’t just an incremental improvement; it’s a paradigm shift. Instead of relying on thousands of labeled samples, these models can classify proteins with just a handful of examples. Imagine accelerating drug discovery, optimizing enzyme engineering, or even uncovering new biological pathways—all with fewer resources and greater accuracy. This article dives into the mechanics of a deep few-shot network for protein family classification, explaining how it works, why it matters, and what it means for the future of bioinformatics.
Let’s break it down.
Understanding Protein Family Classification
Proteins perform countless biological functions, from catalyzing chemical reactions to transporting molecules. Their classification into families helps researchers predict function, understand evolutionary relationships, and identify potential therapeutic targets. However, conventional classification techniques, such as sequence alignment and homology modeling, struggle when faced with low-sample or novel proteins.
Challenges in Protein Family Classification
- Data Scarcity: Many protein families have limited labeled data, making it difficult for traditional models to generalize.
- Computational Complexity: Analyzing vast amounts of protein sequence data requires substantial computational power.
- Structural Variability: Small mutations can lead to significant functional changes, complicating classification efforts.
These challenges underscore the need for a more adaptable and efficient classification approach, which is where deep few-shot learning comes into play.
The Power of Deep Few-Shot Learning in Protein Classification
What is Few-Shot Learning?
Few-shot learning (FSL) is a machine learning technique that enables models to make accurate predictions using minimal training samples. Unlike traditional deep learning models that require thousands of examples, few-shot models can generalize from just a few.
How Few-Shot Learning Works in Protein Classification
- Embedding Proteins into a Feature Space: The model learns a compact representation of protein sequences, capturing essential features.
- Metric-Based Learning: Instead of classifying based on direct comparison, FSL uses distance metrics to compare new proteins against known family representatives.
- Task-Specific Adaptation: The model fine-tunes itself dynamically, improving classification accuracy even with limited data.
By leveraging these principles, deep few-shot networks enhance protein classification accuracy while reducing reliance on large labeled datasets.
Implementing a Deep Few-Shot Network for Protein Family Classification
Key Components of a Few-Shot Network
- Convolutional Neural Networks (CNNs): Extract hierarchical features from protein sequences.
- Recurrent Neural Networks (RNNs) and Transformers: Capture sequential dependencies and contextual relationships.
- Prototypical Networks and Siamese Networks: Optimize learning efficiency by comparing proteins within an embedding space.
Training a Few-Shot Model for Protein Classification
- Data Preparation
- Curate a diverse dataset with representative protein families.
- Preprocess sequences using feature extraction techniques like one-hot encoding and embedding vectors.
- Model Architecture Selection
- Choose between CNNs, RNNs, or transformers based on the complexity of protein structures.
- Integrate attention mechanisms to enhance feature importance.
- Optimization and Fine-Tuning
- Use transfer learning to leverage pre-trained models.
- Employ meta-learning techniques to refine performance on limited samples.
- Evaluation and Validation
- Measure accuracy using precision, recall, and F1-score.
- Validate on independent datasets to ensure generalizability.
Applications of Deep Few-Shot Networks in Bioinformatics
Drug Discovery and Personalized Medicine
Few-shot models accelerate drug development by identifying functional protein targets with minimal experimental data. This enables the discovery of novel therapeutics for rare diseases.
Enzyme Engineering and Industrial Biotechnology
By classifying enzymes efficiently, researchers can design biocatalysts for industrial applications, such as biofuel production and waste degradation.
Evolutionary Biology and Genomic Research
Few-shot learning helps trace evolutionary relationships by classifying newly discovered proteins into ancestral families, shedding light on molecular evolution.
Future Directions and Challenges
Enhancing Model Generalization
Current few-shot networks struggle with highly divergent protein families. Improving generalization through advanced embedding techniques is crucial.
Integrating Multimodal Data
Combining protein sequence, structure, and interaction data could further enhance classification accuracy.
Addressing Interpretability
Deep learning models are often viewed as “black boxes.” Developing explainable AI approaches for protein classification will boost trust and adoption.
Conclusion
Deep few-shot learning is revolutionizing protein family classification. By overcoming data scarcity and improving accuracy, these models pave the way for advancements in bioinformatics, drug discovery, and industrial applications. As research continues, the integration of few-shot learning with multimodal data and explainable AI will further enhance its impact. The future of protein classification is here, and it’s powered by deep learning.